Template Queries¶
Another method of searching CandidaMine is through the use of templates (predefined queries). Popular templates are displayed on the home page, grouped by category (Genes, Protein, Homology, etc.) see Fig. 4. The full list of templates may be viewed by clicking the Templates menu tab. Fig. 5.
Generate query code¶
The code for each query may be obtained by clicking on the arrow next to Generate Python Code and choosing the desired language from the pull-down menu. The language options are Python, Perl, Java, Ruby, JavaScript, and XML.
Download results¶
The search results may also be downloaded by clicking the Export button above the table and choosing the desired format from the pull-down menu to the right of the File name field (blue box in the figure below). Available formats are tab-separated values, comma-separated values, XML, and JSON. When the results contain genomic features, they may also be downloaded in FASTA, GFF3, or BED format. Other options may be specified in the submenu to the left of the download box (orange box in the figure below). By default, all rows and all columns are downloaded, but individual columns may be included or excluded by clicking on the toggles next to the column headers in the All Columns submenu. The number of rows and row offset are set in the All Rows submenu. Download the results as a compressed file by choosing GZIP or ZIP format in the Compression submenu (default is No Compression). Column headers are not added by default but may be included under the Column Headers submenu. Finally, the Preview submenu displays the first three rows of the file to be downloaded so that the desired format and options may be finalized before beginning the download. When ready, click the Download file button to download the results.
Customize output¶
Click the Manage Columns button to customize the results table layout. Edit or remove active filters by clicking the Manage Filters button. Click Manage Relationships to specify the entity relationships within the query.
Optional filters¶
Some templates have optional filters that are disabled by default. For example, the GO Term –> Gene template has an additional filter for specifying a GO evidence code. To enable this filter, click ON below GO Evidence Code > Code.
Examples¶
Genes to Proteins¶
INDELS in coding regions¶
To get all insertion and deletions in coding regions you can run Insertions/Deletions in CDS region template. The templates has some filters to constrains the search for organism of interset , specfic gene, and optionals strains and study PMID as shown in Fig. 9.
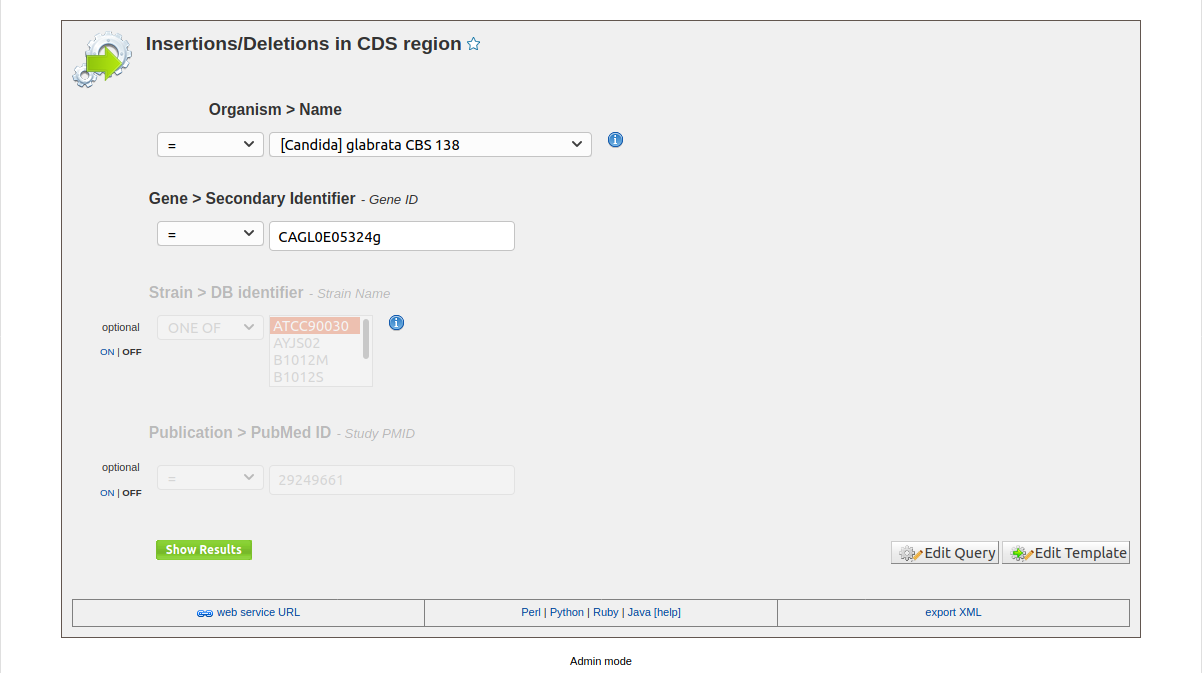
Fig. 9 Insertions/Deletions in CDS region Template Query




